Intro to RNA-seq
Welcome All!
Dates
- Day 1: 10:00am - 4pm Tuesday 4th July
- Day 2: 10:00am - 4pm Friday 7th July
Pre-requisties:
Proficient in:
- R
- Command line
- HPC
All the courses will assume prior knowledge from Intro to R and Intro to Command-line/HPC
Before the course please make sure you have:
- Charger
- Positive attitude!
- RStudio and R installed (see the instructions below)
- A login to the wolfpack
- See the instructions below and also instructions from the intranet.
- If you can
sshin by running the command below
- Optional: Code Editor, see below
At the end of the workshop you will be able to:
- Be able to process data from FASTQ to count matrix using the HPC
- Transfer results from the HPC to your local laptop
- Using DESeq for differential expression for a basic experiment
You will not be taught:
- How to apply the RNA-seq pipeline in batch
- Complicated analysis using DESeq2
DSP Helpdesk
To ask help from Derrick and select the portal or email dsphelp@garvan.org.au
Installing Code Editor
We would also recommend installing a code editor. Our team will be using vim to write scripts. However, Visual Studio Codes is a better, less error-prone method of writing code. Feel free to use your editor of choice. You can install Visual Studio Codes from https://code.visualstudio.com/.
Accessing the Wolfpack, Garvan’s HPC
- For Mac users can use the Terminal program. You can open it by spotlight-searching “Terminal.” Alternatively, you can use iTerm2, which is a macOS terminal replacement that I personally prefer.
- Windows users:
- First, check if you have the Command Prompt or PowerShell program locally. You might need to enable SSH using the tutorial recommended by John Reeves: How to Enable and Use Windows 10’s Built-in SSH Commands..
- Second, if you do not have either Command Prompt or PowerShell installed, it is likely your laptop has a Windows OS before 10. Therefore, I recommend installing PuTTY which is an open source software.
Logging on
You log on to the server using your username and a program that lets you connect via a “secure shell (SSH)”. If you use a Mac, you simply need to open the Terminal. Terminal is generally found in the “Other” folder in Launchpad, or just search for “Terminal” with Spotlight. Once open, Keep in Dock for handy future access. If using Windows, either open PowerShell or PuTTy as mentioned previously.
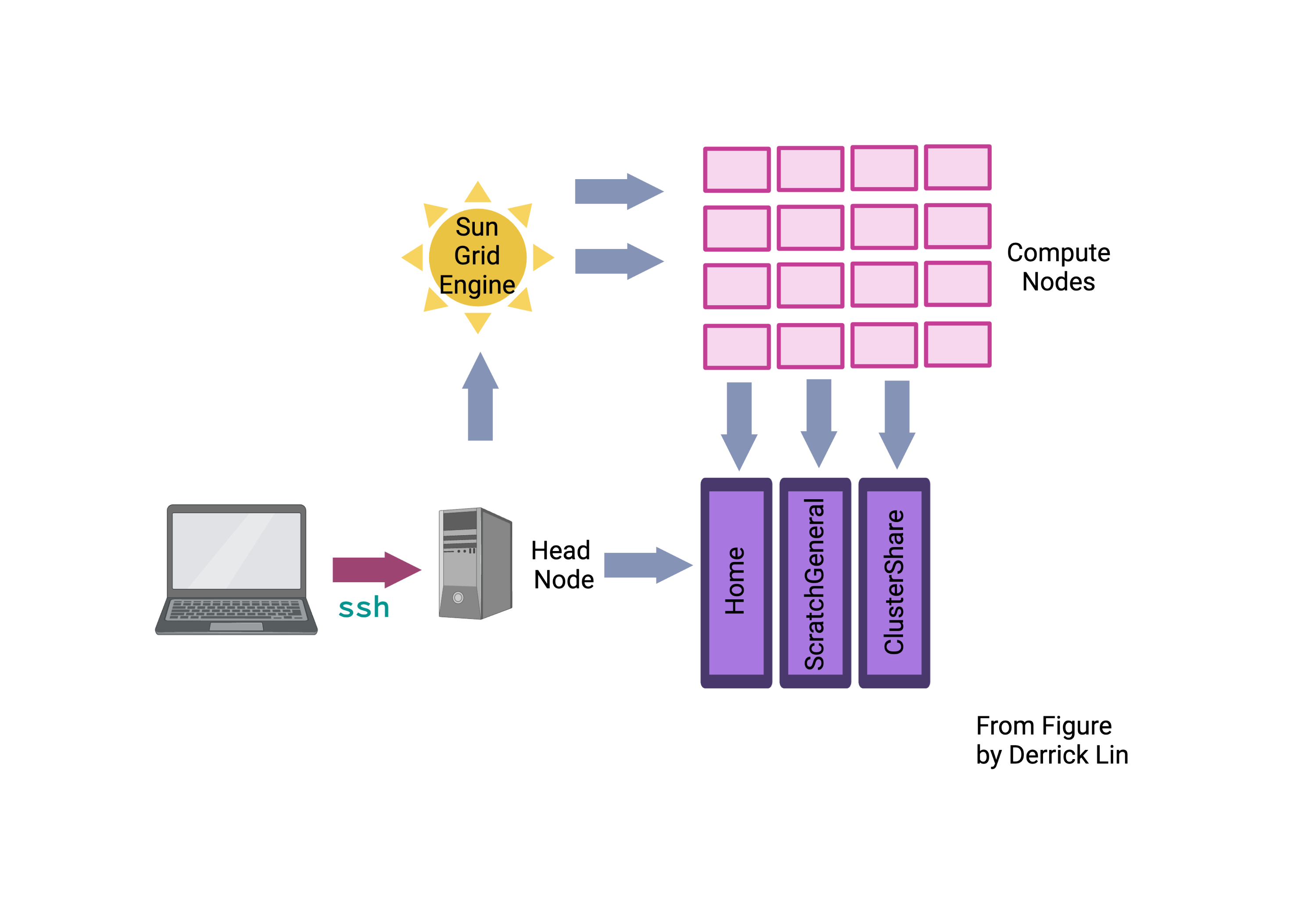 Above is a schematic that displays the setup of the Wolfpack. We will explain complicated part of the diagram concerning volumes and compute nodes in future sessions. What we are doing is the first pink arrow, log in into the login nodes dice01.garvan.unsw.edu.au or dice02.garvan.unsw.edu.au.
Above is a schematic that displays the setup of the Wolfpack. We will explain complicated part of the diagram concerning volumes and compute nodes in future sessions. What we are doing is the first pink arrow, log in into the login nodes dice01.garvan.unsw.edu.au or dice02.garvan.unsw.edu.au.
To log on from Mac OSX (or a UNIX machine), open the Terminal and type at the prompt (replacing username with your own username ):
$ ssh username@dice01.garvan.unsw.edu.au
Change the username to your personal login id, or dice01 to dice02 for different login nodes.
Logging on from outside the Garvan. Remember that to log on from outside Garvan, you will need to connect to the virtual private network (VPN).
RStudio and R installation
R and RStudio are separate downloads and installations. R is the underlying statistical computing environment, but using R alone is no fun. RStudio is a graphical integrated development environment (IDE) that makes using R much easier and more interactive. You need to install R before you install RStudio. Please choose the operating system (OS) that you use.
Garvan Owned Devices
If anyone has a Garvan-owned Windows device and requires any software installed, they need to contact ithelp@garvan.org.au to get it done (as they might not have administrator privileges) and if they have Garvan-owned MacBook, they can install any software by themselves
Windows
To install R and RStudio
- Download R from the CRAN website.
- Click on and run the .exe file that was just downloaded. Leave all default settings in the installation options.
- Go to the RStudio download page
- Under Installers select RStudio x.yy.zzz - Windows XP/Vista/7/8 (where x, y, and z represent version numbers)
- Double-click the file to install it
- Once it’s installed, open RStudio to make sure it works and you don’t get any error messages
If you have an error do not stress. Let us know at the beginning of the session, we will be able to help out.
macOS
To install R and RStudio
- Download R from the CRAN website.
- Note that for older Macs, without the M1/M2 chip, you must select the appropriate Intel download from the CRAN homepage.
- Click on the pkg file that was just downloaded. Leave all default settings in the installation options.
- Go to the RStudio download page
- Under Installers select RStudio x.yy.zzz - Mac OS X 10.6+ (64-bit) (where x, y, and z represent version numbers)
- Double-click the file to install it
- Once it’s installed, open RStudio to make sure it works and you don’t get any error messages
If you have an error do not stress. Let us know at the beginning of the session, we will be able to help out.